伊人直播
Single-cell analysis not only reveals the real physiological state of cells, but also enables non-discriminatory analysis of rare cell populations, which is crucial for a comprehensive understanding of the classification of cell populations. Over the past five years, Professor Bai’s group has made a series of progress in single-cell omics research. and realized multiplexed protein detection of trace biological samples by designing signal amplification strategies based on mass tag labelling (J. Am. Chem. Soc. 2019, 141, 72). A multi-dimensional organic mass cytometry platform was developed to realize simultaneous analysis of multi-target proteins and more than 100 metabolites within a single cell, providing new insights for the study of tumor drug resistance mechanisms (Angew. Chem. In. Ed. 2021, 60, 1806). Recently, Bai’s group reviewed and prospected the research progress in the field of single cell omics analysis based on mass spectrometry (TrAC, Trends Anal. Chem. 2023, 164, 117086).
Single-cell metabolomics analysis can systematically "read" the physiological and functional states of cells and is essential for revealing cell phenotypic heterogeneity and elucidating the molecular mechanisms of biological processes. However, due to the limited identification ability of existing single-cell metabolomics methods, the short online analysis time of single cells leads to the inability to obtain MS/MS information as well as distinguish isomers. Addressing these challenges, the Bai Yu research group from the College of Chemistry and Molecular Engineering at Peking University developed an in-depth organic mass cytometry platform, which can realize online and efficient cell lysis and high-resolution mass spectrometry analysis. The analysis time of single cells is 40 times longer than the existing technology, and abundant MS/MS spectra for metabolite identification can be obtained. The platform identified up to 600 metabolites per single cell, enabling comprehensive qualitative and quantitative analysis of single cell isomers (Fig. 1), and revealing the existence of fine subtypes of MCF-7 breast cancer cells. On May 23, 2024, the research results titled "In-depth organic mass cytometry reveals differential contents of 3-hydroxybutanoic acid at the single-cell level" were published in the journal Nature Communications (Nature Communications, 2024, 15, 4387).
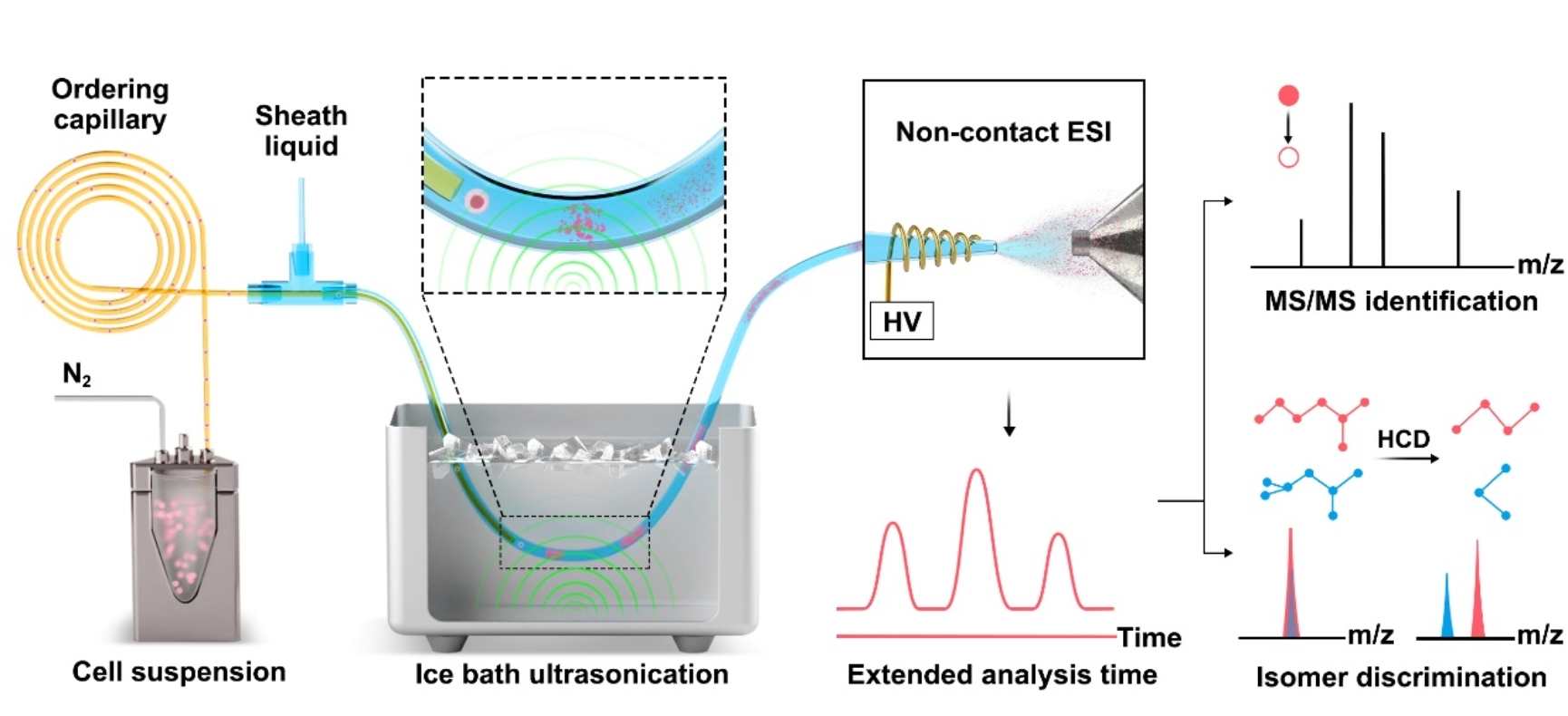
Fig. 1 The workflow of in-depth organic mass cytometry.
The in-depth organic mass cytometry platform consists of an on-line high-efficiency single-cell lysis device and non-contact electrospray high resolution mass spectrometry analysis. The cell suspension was pumped into the inner ordering capillary in which single-cell sorting was achieved via Dean flow. The sheath fluid capillary is coaxially connected with the separation capillary through a T-value. Low-energy ultrasonication was employed to realize online cell lysis after mixing the cell solution with sheath fluid. Notably, ice bath conditions were used to prevent metabolite degeneration caused by the heating effect from continuous ultrasonication. With continuous single cell injection, an average analysis time of about 24 seconds per single cell can be obtained, which can effectively acquire high-resolution MS and MS/MS information. The substantial MS/MS data also enables the identification and relative quantification of isomers (Fig. 2).
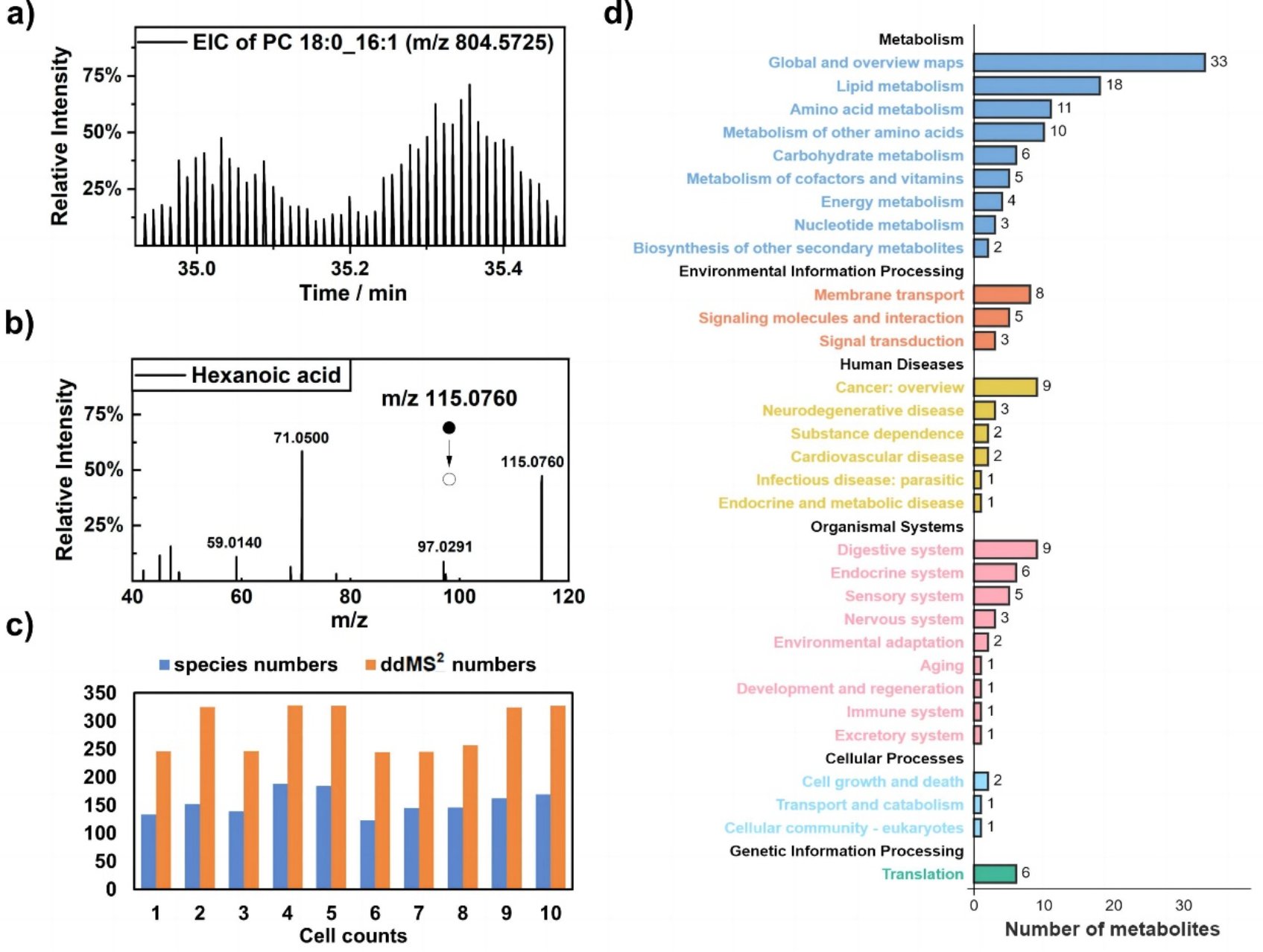
Fig. 2 Profiling of metabolites according to MS2 at the single-cell level.
Based on the metabolite information of different types of tumor cells, the platform demonstrates the universal cell typing capabilities. Using the relative content of 3-hydroxybutyric acid/4-hydroxybutyric acid isomers, three subtypes of MCF-7 cells were successfully distinguished by UMAP visualization method. Combined with the 10×Genomics single cell sequencing analysis, western blot assay and fluorescence activated cell sorting assay, this study confirmed that the abundance difference of 3-hydroxybutyric acid was the main reason of tumor cell subtyping from the transcriptomic level and protein expression level, and 3-hydroxybutyric acid was positively correlated with the expression levels of various downstream anti-oxidative stress target proteins, such as, metallothionein MT2A and heterogeneous ribonucleoprotein hnRNP A1. These findings suggest that variations in 3-hydroxybutyric acid abundance may confer different abilities on tumor cells to resist oxidative stress environments. This work observed potential cell subtypes of MCF-7 cells for the first time, offering a new approach to understanding tumor heterogeneity and advancing precise therapy for breast cancer.
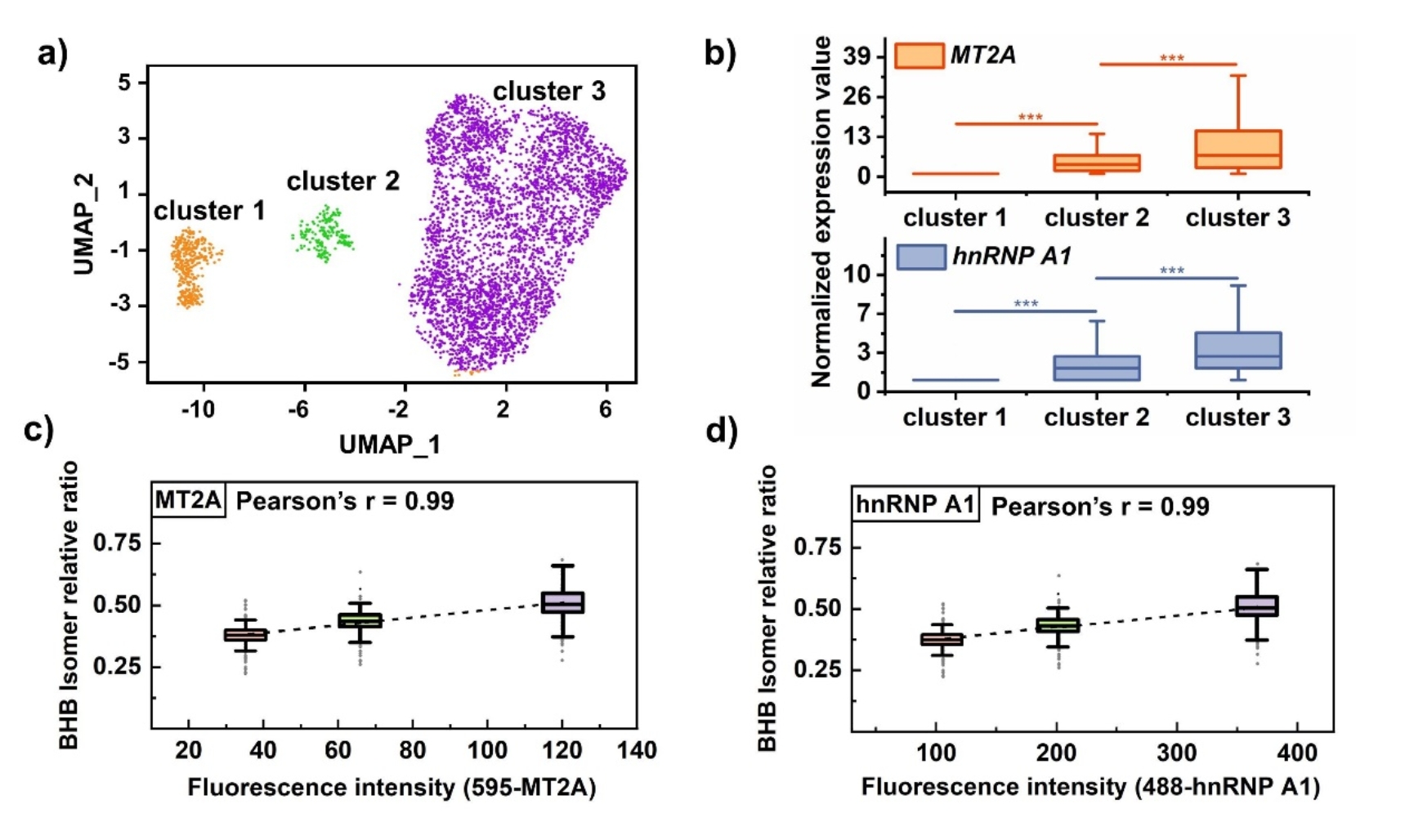
Fig. 3 Clustering of MCF-7 cells with BHB/GHB isomer information.
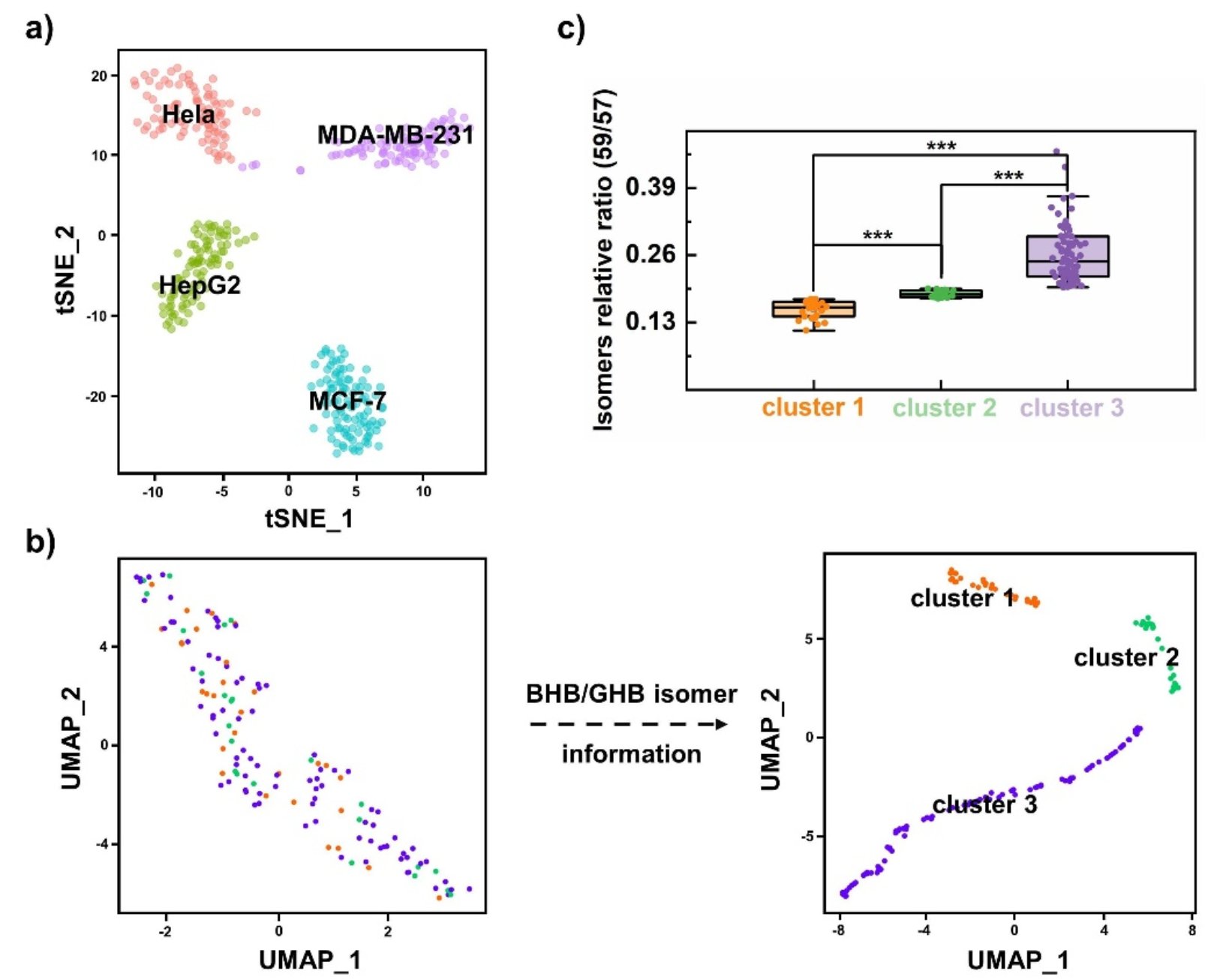
Fig. 4 10x Genomics single-cell transcriptome sequencing verification.
Prof. Yu Bai from the College of Chemistry and Molecular Engineering at Peking University is the corresponding author. Mr. Shaojie Qin (Ph.D. candidate) from the College of Chemistry and Molecular Engineering at Peking University is first author of the article. The research was funded by the National Natural Science Foundation of China, the Ministry of Science and Technology, and the Beijing National Laboratory for Molecular Sciences. The sequencing data analysis was supported by Associated researcher Lu Wen, Jiansen Lu and Hongmin Yu from the Biomedical Frontier Innovation Center of Peking University. The research was also supported by Dong Liu, a teacher from the Mass Spectrometry Platform of the National Protein Science Research (Beijing) Branch Center of Peking University.
Link to the original article: //www.nature.com/articles/s41467-024-48865-2