伊人直播
When plants encounter stress, a molecular revolution is triggered at all levels, from gene transcription to protein regulation. Abiotic stresses, such as salt stress, significantly impair crop growth and productivity. At the epigenetic level, RNA modifications, particularly N6-methyladenosine (m6A), have been shown to mediate several processes, including plant growth, development, and responses to external or internal stresses. Upon salt stress, there is a dynamic change in m6A level. However, the precise mechanism underlying this phenomenon and its full implications remain unclear.
Recently, the research group led by Professor Guifang Jia from the College of Chemistry and Molecular Engineering at Peking University and the Peking-Tsinghua Center for Life Sciences published a study in The Plant Cell entitled "The m6A reader ECT8 is an abiotic stress sensor that accelerates mRNA decay in Arabidopsis". This research elucidated how ECT8 functions as a salt stress sensor by binding to m6A-modified mRNA. ECT8 interacts directly with Decapping 5 (DCP5) within the processing body (P-body), thereby participating in the 5' end decapping pathway. This mechanism helps maintain physiological homeostasis in plants under stress conditions.
In this study, the authors first demonstrated that ECT8 is an m6A reader in Arabidopsis (Fig. 1). Analysis of ECT8 expression levels in databases showed a significant increase under various abiotic stress stimuli (e.g., salt stress, oxidative stress, drought stress) compared to m6A methyltransferases, demethylases, and other YTH family proteins.
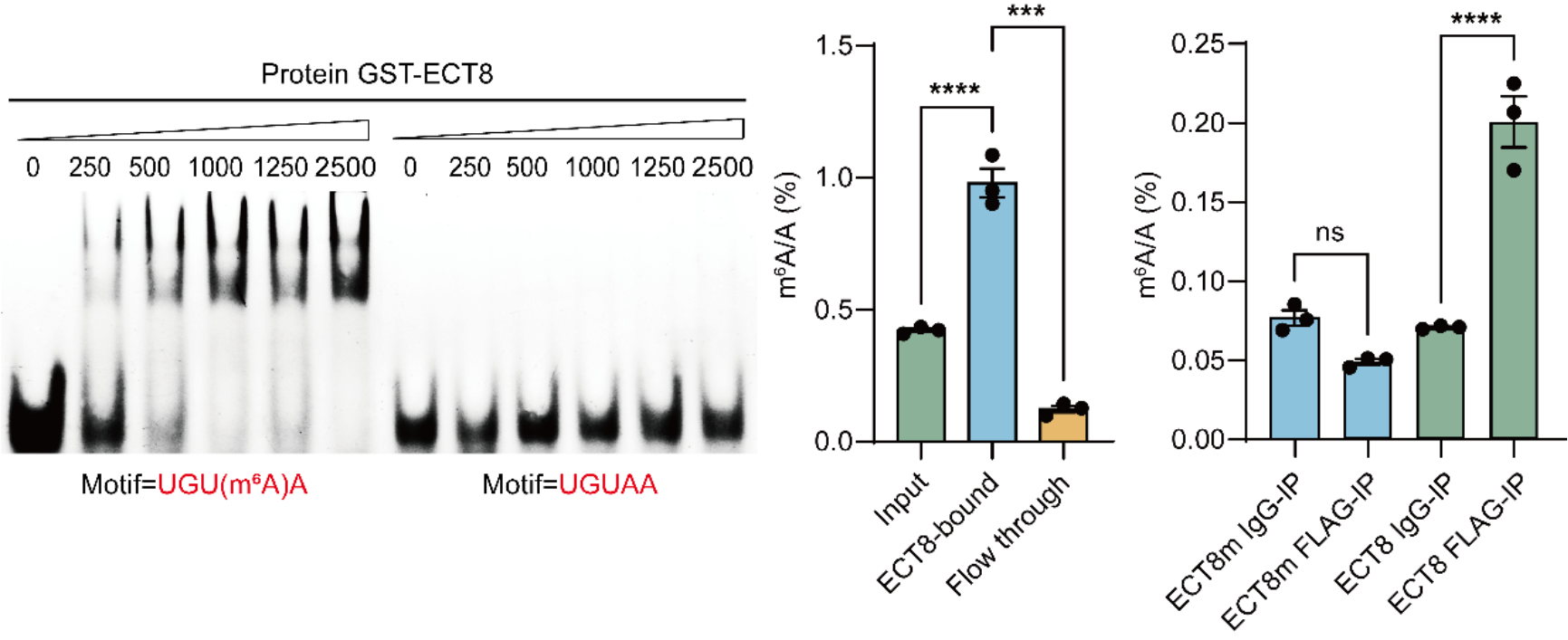
Fig. 1 ECT8 is an m6A binding protein in Arabidopsis.
Following this, the authors used salt stress as an example, employing nuclear run-on and protein immunoblotting to prove that both transcripts and protein levels of ECT8 significantly rise under stress conditions. Subsequent results showed that the absence of ECT8 or the loss of its m6A binding function resulted in delayed germination, and shortened root length under salt stress (Fig. 2). These results suggest that ECT8 responds rapidly to external stimuli and functions to maintain plant physiological homeostasis under stress conditions.
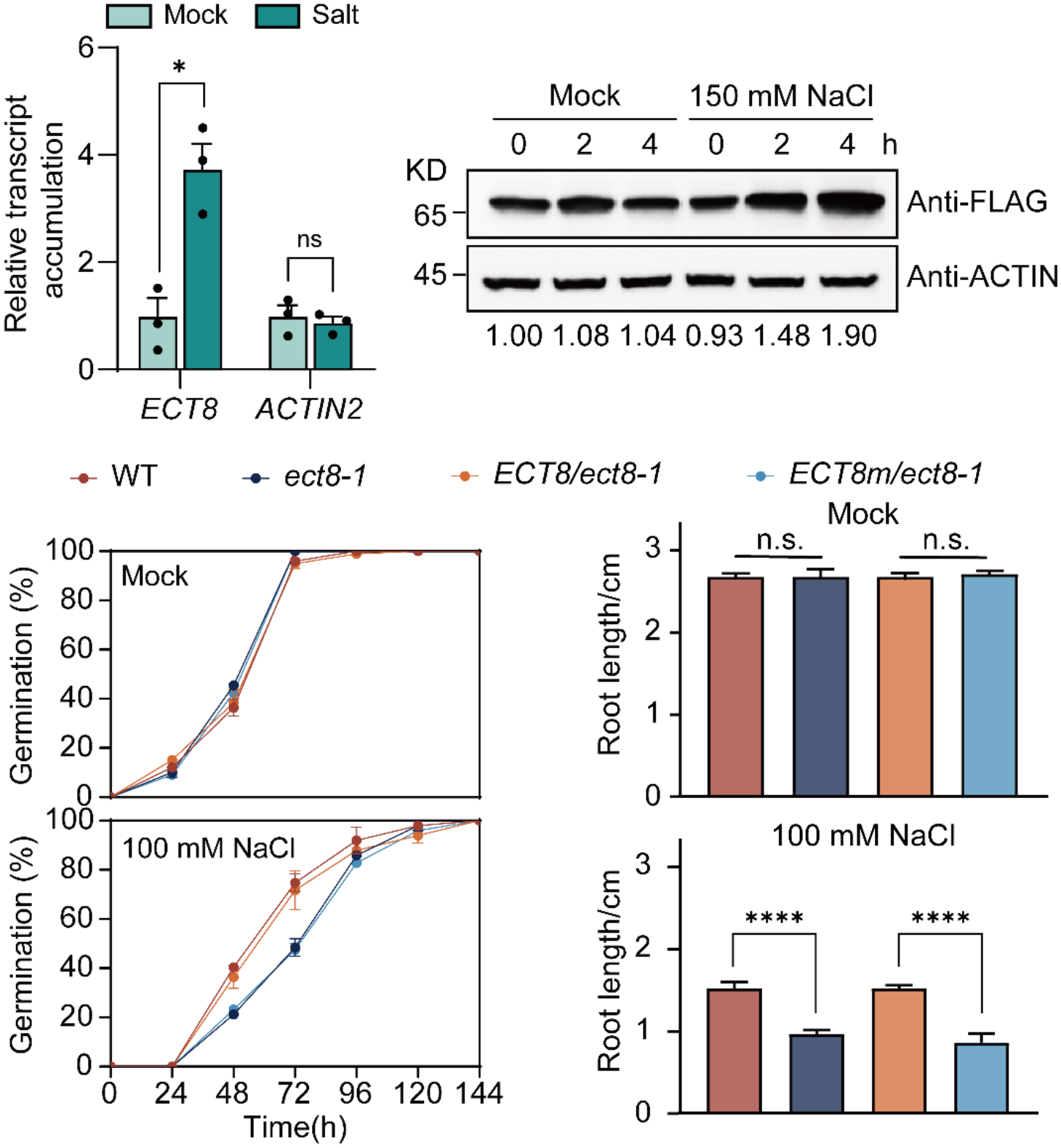
Fig. 2 ECT8 is required for salt stress response in an m6A-dependent manner.
To explore the specific function of the ECT8 protein, the authors used MeRIP-Seq, FA-CLIP, and RNA life-time sequencing to find that salt stress increases ECT8's binding ability to the m6A region of target transcripts. Furthermore,, the absence of ECT8 led to increased expression and prolonged half-life of these transcripts (Fig. 3).
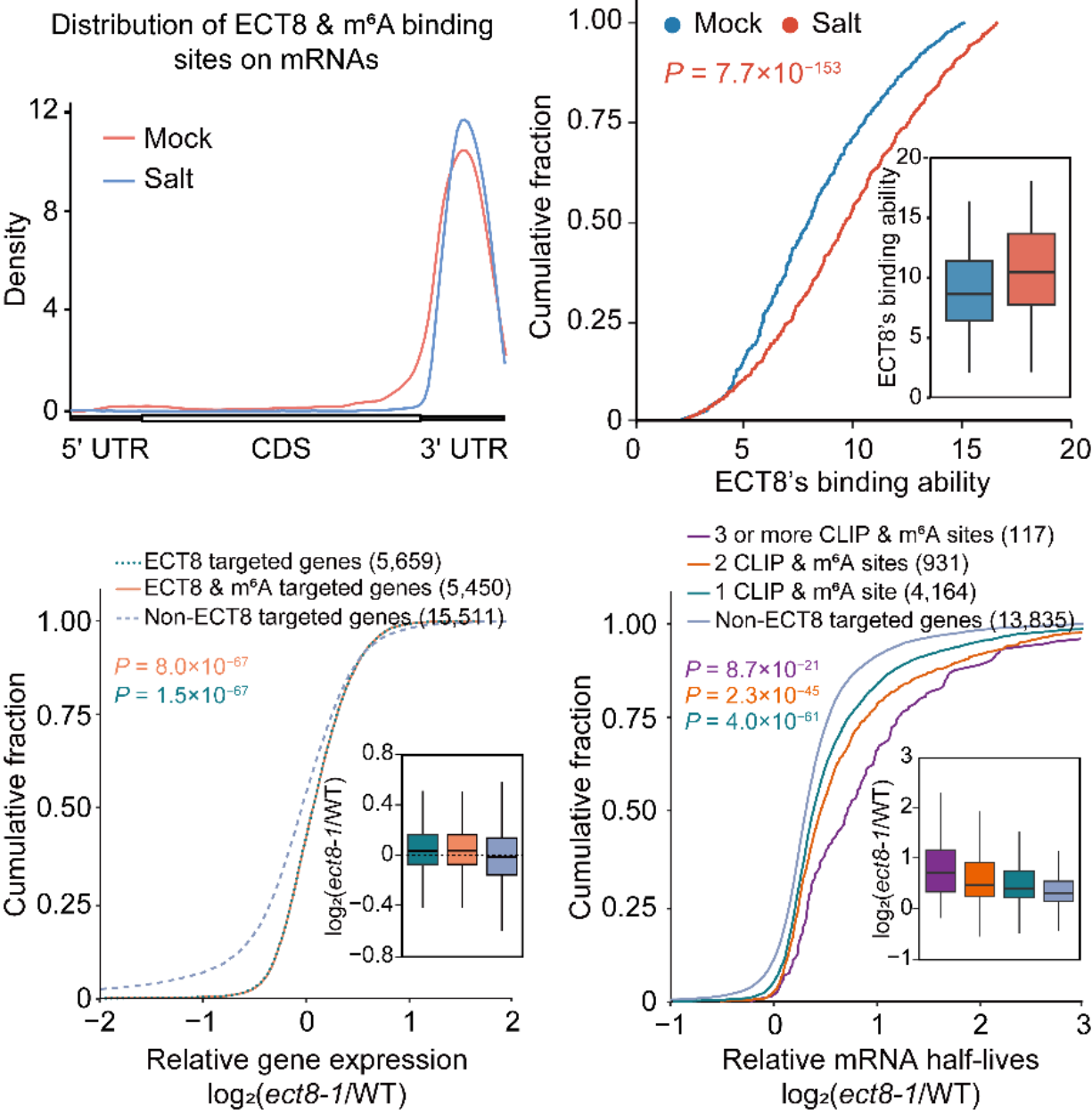
Fig. 3 ECT8 binds to mRNA 3' UTR regions and facilitates the degradation of m6A-modified mRNAs.
Considering the presence of a disordered prion-like domain in the ECT8 protein structure, the authors hypothesized and confirmed that ECT8 accelerates the degradation of its target transcripts through direct interaction with the decapping protein DCP5 within P-bodies. Their research indicated that ECT8 binds to m6A-modified mRNAs, thereby accelerating their degradation, especially those of negative regulators of salt stress responses (Fig. 4). The results demonstrated that ECT8 acts as an abiotic stress sensor, facilitating mRNA decay, vital for maintaining transcriptome homeostasis and enhancing stress tolerance in plants. Importantly, this study advances our understanding of epitranscriptomic gene regulation and presents potential applications for breeding more resilient crops in rapidly changing environmental conditions.

Fig. 4 ECT8 accelerates the degradation of salt stress negative regulators through direct interaction with DCP5 within P-bodies.
Zhihe Cai (a Ph.D. candidate), Qian Tang (a Ph.D. graduate) and Peizhe Song (a postdoc) from the College of Chemistry and Molecular Engineering, Peking University are co-first authors of the article. Prof. Guifang Jia from the College of Chemistry and Molecular Engineering and the Peking-Tsinghua Center for Life Sciences at Peking University is the corresponding author. The research was supported by the National Natural Science Foundation of China, the National Key R&D Program of China and the Beijing Natural Science Foundation, Beijing National Laboratory for Molecular Sciences.
Link to the original article: //academic.oup.com/plcell/advance-article/doi/10.1093/plcell/koae149/7687916?searchresult=1