伊人直播
Biomolecules in solution undergo rapid conformational changes driven by thermal fluctuation with a rugged energy landscape. Traditional ensemble averaging methods face challenges capturing these fleeting, high-energy intermediate states. The advances in single-molecule biophysical techniques, such as single-molecule fluorescence, nanopore detection, and single-molecule force spectroscopy, have greatly improved temporal and spatial resolution, enabling observation of biomolecular dynamics at relevant scales. Cryo-electron microscopy (cryo-EM) with three-dimensional reconstruction algorithms like CryoDRGN has enabled the reconstruction of biomolecular structures from latent spaces, while time-resolved cryo-EM (trEM) provides insights into the temporal evolution of biomolecular 3D structures of during interactions. However, directly imaging and analyzing the dynamic processes of biomolecules in solution remains challenging.
Single-molecule liquid-phase transmission electron microscopy (LP-EM) excelsin resolving the structural dynamics of biomolecules during interactions with millisecond temporal resolution. With image analysis methods based on deep learning algorithms and rigorous evaluation of electron beam effects, LP-EM can reveal transient dynamics and intermediate conformational states in biomolecular systems such as proteins and DNA/RNA.
Recently, Prof. Huan Wangʼs research group from the College of Chemistry and Molecular Engineering at Peking University was invited to publish a "Forum" article titled "Imaging Biomacromolecules in Action with Liquid-phase Electron Microscopy" in Trends in Chemistry. The article explores the application of LP-EM in analyzing the dynamic processes and intermediate states of biomolecules in solution, along with advanced image analysis and 3D reconstruction algorithms.
Professor Wangʼs group is dedicated to advancing LP-EM imaging techniques and dynamic quantitative analysis methods. Their pioneering research has been featured in several prominent journals, including Adv. Mater. 2017, 29, 1703555, ACS Nano 2018, 12, 8572, Proc. Natl. Acad. Sci. U.S.A. 2019, 117, 201916065, Microsc. Microanal.2021, 27, 3, ACS Nano 2022, 11, 18526, Chem Commun. 2023, 59, 1701, Proc. Natl. Acad. Sci. U.S.A. 2024, 121, e2314797121; Chin. J. Chem. 2023, 41, 679; Acta Polymerica Sinica 2024, 55(1), 129-148. The innovative use of LP-EM has led to groundbreaking studies on the conformational distribution of circular DNA, the dynamic assembly of liposomes, the hybridization and assembly process of single-stranded DNA (ssDNA), and the enzyme-catalyzed growth process of double-stranded DNA (dsDNA). This research propels the development of LP-EM technology and provides new perspectives and methods for analyzing the dynamic processes of biomolecules in solution.
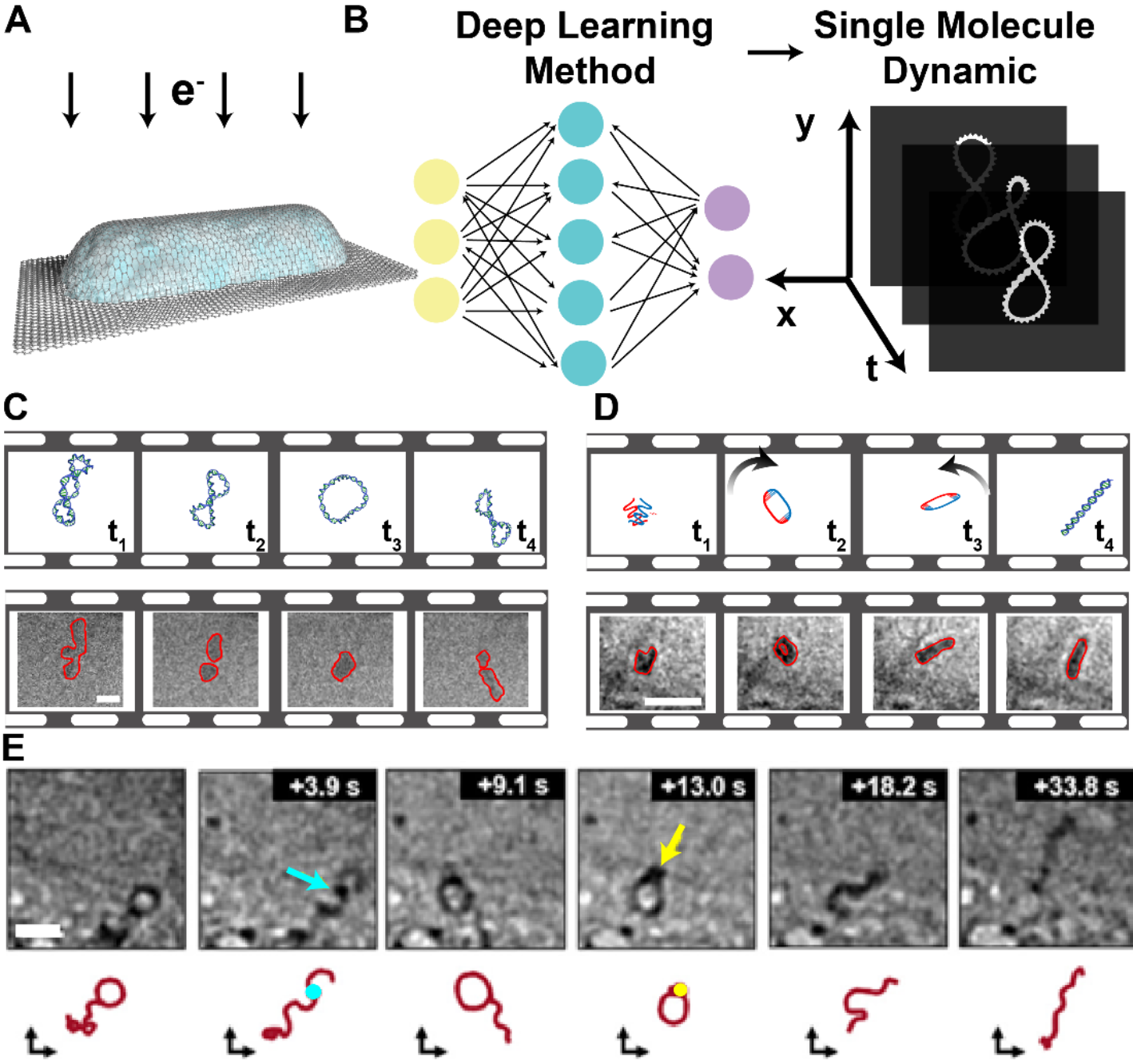
Fig. 1 LP-EM imaging of individual biomacromolecules and their interaction. A. Schematic diagram of a typical GLC. B. Schematic diagram of single-molecule analysis through deep-learning-based methods for movies obtained by LP-EM. C. Equilibrium system: reversible transitions between conformational states of a circular DNA molecule. States are characterized as different loop numbers from 3-loop (t1), 2-loop (t2) to 1-loop (t3), and back to 2-loop (t4). Scale bar: 10 nm. Images adapted from reference 8 with permission. D. Interaction system: pathway and intermediate loop state (t2) resolved from time-lapsed LP-EM images during hybridization of ssDNA (t1-t3) forming dsDNA (t4). Scale bar: 30nm. Images adapted from reference 9 with permission. E. Enzyme system: DNA (red) polymerization by enzymes of Bgl1 (cyan at 3.9s) and T4 ligase (yellow at 13.0s). Scale bar: 20nm. Images adapted from reference 4 with permission.
In the near future, with continued advancements inneural network-based image analysis algorithms, these methods are anticipated to enable imaging of individual protein molecules and analysis of their transient dynamics. This progress will resolvehigh-energy, rare conformational states that differ from crystal structures and facilitate quantitative analysis of their conformational distributions, allowing comparisons of changes during interactions. Additionally, the development of 3D reconstruction algorithms is expected to reconstruct the time-resolved 3D structures of biomolecules from 2D projections in LP-EM movies. This will provide more detailed information on conformational changes, shedding light on fundamental biophysical questions related to biomolecular functions, such as the mechanisms of conformational selection,induced fit in biological interactions, and disease formation processes.
This work was supported by the National Natural Science Foundation of China, the National Biomedical Imaging Center, the Key Laboratory of Polymer Chemistry & Physics, Peking University, and the Spectroscopy Center of Peking University. The corresponding author is Assistant Professor Huan Wang from the College of Chemistry and Molecular Engineering at Peking University, and the first author is Ph.D.student Jiaye Li from the College of Chemistry and Molecular Engineering, Peking University. Collaborators include Professor He Sun from the College of Future Technology at Peking University.
Link to the original article: //doi.org/10.1016/j.trechm.2024.04.004